About me
I am passionate about leveraging data analytics and machine learning to tackle complex problems related to human health.
I have a BS and PhD in Physics / Computational Biology and completed a postdoc in structural biology. My academic work focused on high-performance computation, simulation, and cryoEM to elucidate the assembly mechanism of bacterial membrane proteins, aiding in the development of novel antibiotics. This research resulted in publications in Nature Communications (2021) and PNAS (2018), where I identified specific protein mechanisms relevant to antimicrobial resistance.
In my professional journey, I’ve sought out experiences that draw upon my scientific expertise while embracing modern AI/ML methods to positively impact human health. After completing a Fellowship at the NYC Data Science Academy, I began a role at Calyxt where I developed analytic tools to help researchers identify metabolic pathways in plants and optimize rare-compound production for pharmaceutical manufacturing.
At EMD Serono (Merck group), I developed algorithms leveraging ML models and NLP data from over 30 million publications, clinical trials, and patents to improve drug target prioritization for immune disorders. I also built custom web applications for visualizing gene “trendiness” scores to enable efficient drug-target prioritization.
Currently, I work as a Scientific Consultant at Metapages (Astera Institute), where I’m helping to develop an interactive computational biology platform that enables the easy creation and sharing of interactive scientific workflows.
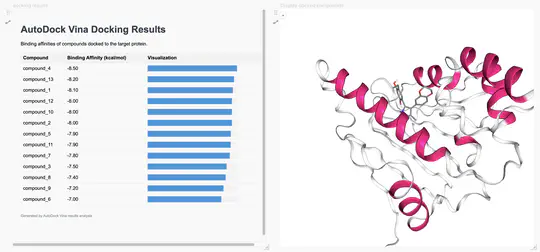
My technical expertise includes Python (numpy, pandas, scikit-learn, pytorch, tensorflow, biopython, dash, plotly), R (DESeq2, Bioconductor, rshiny, dplyr, tidyr, ggplot2), ML/AI methods, cloud computing, molecular docking, protein modeling, and molecular dynamics simulations.
- Molecular Dynamics Simulation
- Drug Discovery
- Computational Protein Design
PhD in Physics and Chemistry, 2019
Georgia Institute of Technology
MS in Physics, 2015
Georgia Institute of Technology
BS in Physics and Mathematics, 2012
University of Michigan
Projects
Experience
- Developing an interactive computational biology platform that enables easy creation and sharing of scientific workflows
- Implementing containerized computational environments for reproducible research
- Creating modular analysis components for genomics and protein structure analysis
- Applying deep learning approaches to protein design challenges
- Developed algorithms leveraging ML models to analyze over 30 million publications, clinical trials, and patents
- Created text mining and time-series analysis pipelines to identify high-interest genes for drug target prioritization
- Built custom web applications for visualizing gene “trendiness” scores to enable efficient decision-making
- Implemented NLP methods to extract and analyze drug target information from scientific literature
- Developed ML models to predict rearrangement of genetic elements that maximize production of rare compounds in plants
- Created analytic tools to help researchers identify metabolic pathways in plants for pharmaceutical manufacturing
- Implemented a data processing pipeline for analyzing plant metabolomics datasets
- Collaborated with research teams to optimize experimental designs and analyze complex biological data
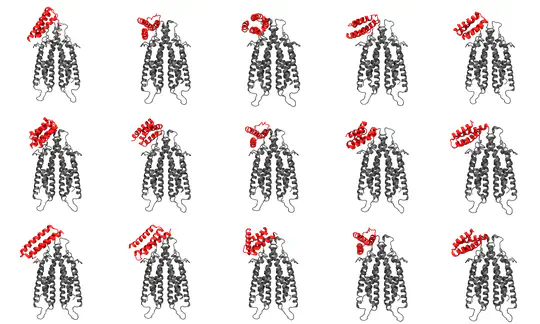
- Elucidated assembly mechanisms of bacterial membrane proteins using cryo-electron microscopy and molecular dynamics
- Published findings in Nature Communications (2021) and PNAS (2018) on protein mechanisms relevant to antimicrobial resistance
- Developed high-performance computational simulations to study membrane protein-lipid interactions
- Identified potential targets for novel antibiotic development through structural and functional analysis
- Conducted research on bacterial outer membrane protein assembly using computational methods
- Developed novel approaches to analyze protein-lipid interactions in complex membrane environments
- Performed large-scale molecular dynamics simulations on high-performance computing clusters
- Published multiple peer-reviewed papers on membrane protein folding and assembly