CD20 Protein Binder Design
Project Overview
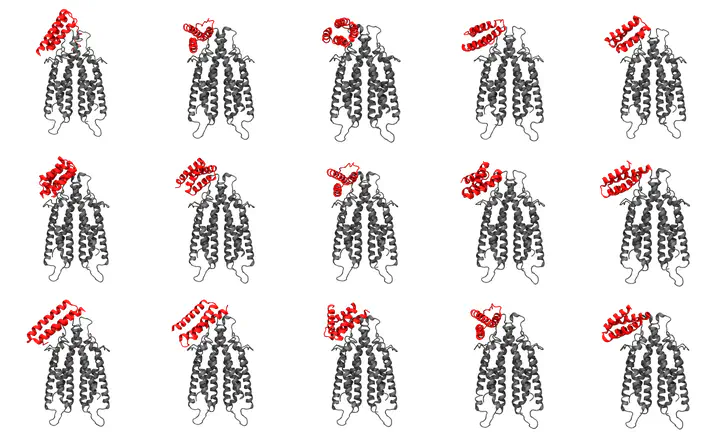
In this project, our team developed a computational pipeline to design novel protein binders specifically targeting CD20, a protein found on the surface of B cells that is a key target for various immunotherapies. This work demonstrates the application of cutting-edge deep learning approaches and molecular simulation techniques to create functional proteins with potential therapeutic applications.
The Challenge
Designing proteins that can specifically bind to targets like CD20 represents a significant frontier in therapeutic development. Traditional methods rely on experimental screening of vast libraries—a process that consumes extensive time and resources. Our team of four sought to address this limitation by developing a computational approach that could dramatically accelerate protein binder design, leveraging recent advances in AI and molecular simulation techniques.
Our project focused on CD20, a critical protein found on B-cell surfaces and a key target for numerous immunotherapies. We aimed to create a systematic pipeline that could design novel protein binders with high specificity and affinity for this therapeutic target.
Our Approach
We began by implementing RFDiffusion to generate initial protein binder structures complementary to the CD20 target region. This phase involved extensive parameter exploration to optimize the binding interface. We tested different configurations, including hotspot-focused designs targeting specific CD20 residues (168-175), broader interface designs covering larger regions (residues 46-210), and beta-model variants with enhanced structural constraints.
For sequence optimization, we applied ProteinMPNN, a deep learning model designed specifically for protein sequence generation. Working with Rosetta FastRelax for structural refinement, we optimized each design for both stable protein folding and strong complementarity with the CD20 extracellular domain.
Structure prediction served as our critical validation step. Using AlphaFold2, we predicted the 3D structures of our designed sequences and implemented filtering criteria to eliminate designs with poor confidence scores or structures that would protrude into the cell membrane when properly aligned with CD20. These practical constraints ensured that our designs would be functional in a biological context.
Refinement Process
Assessing binding stability emerged as one of the most nuanced aspects of our project. We developed a workflow incorporating energy minimization and molecular dynamics simulations for side-chain relaxation. This approach allowed us to evaluate binding stability through multiple metrics: total energy calculations, non-bonded interaction energy between binder and CD20, and binding free energy estimation using Prodigy.
Our process evolved into a multi-round design strategy that balanced exploration and exploitation. We carried forward the top-performing designs through progressive iterations while continuously generating new variants to maintain diversity. Designs meeting our criteria for energy thresholds and RMSD advanced through rounds, each iteration improving stability and affinity. This iterative approach allowed us to effectively navigate the vast protein design space, systematically improving our binders’ qualities while maintaining a computationally feasible search.
Technical Implementation
The success of our pipeline relied on thoughtful computational infrastructure. We used Docker for containerization to ensure reproducibility across different computing environments. GPU acceleration proved essential for the computationally intensive tasks of structure prediction and molecular dynamics simulations. We developed shell scripts to orchestrate the multi-stage process, allowing for minimal human intervention between stages.
For analysis and visualization, we created interactive Jupyter notebooks that enabled our team to examine structural details and binding interfaces. This facilitated collaborative decision-making about which designs to advance in subsequent rounds.
Results and Impact
Our computational design process yielded a collection of high-affinity protein binders specifically targeting the extracellular region of CD20. These final designs demonstrated strong predicted binding affinity, structural stability, and specificity for the target epitope while minimizing interaction with the cell membrane.
The methodology we developed has broader implications beyond CD20. It provides a framework that could be adapted to design binders for other disease-relevant proteins, potentially leading to new therapeutic candidates for conditions where targeted protein binding is beneficial, such as:
- Cancer immunotherapies
- Autoimmune disorders
- Inflammatory diseases
- Infectious disease treatments
By consolidating and re-ranking the top binders from all rounds, our approach ensures that the most promising candidates are identified for potential experimental validation.
Technical Skills Demonstrated
Through this project, our team applied and strengthened expertise in:
- Computational Biology: Protein design, molecular dynamics, binding energy calculations
- Machine Learning: Application of deep learning models for protein structure and sequence prediction
- Software Engineering: Pipeline development, containerization, workflow automation
- Scientific Computing: Data analysis, visualization, and interpretation
Technical Elements
Our pipeline integrated several state-of-the-art computational tools:
- RFDiffusion for de novo protein structure generation
- ProteinMPNN for deep learning-based protein sequence design
- AlphaFold2 for protein structure prediction
- Rosetta for energy calculations and structural refinement
- OpenMM for molecular dynamics simulations
- Prodigy for binding affinity prediction
- Docker for environment containerization
- Python for data processing and analysis
- Jupyter for interactive visualization
This project demonstrates how computational approaches can transform therapeutic protein design, potentially reducing the time and resources required to develop new targeted therapies while expanding the range of addressable targets.
References
- Watson, J.L., Juergens, D., Bennett, N.R. et al. (2023). De novo design of protein structure and function with RFdiffusion. Nature, 620, 1089–1100.
- Bennett, N.R., Coventry, B., Goreshnik, I. et al. (2023). Improving de novo protein binder design with deep learning. Nat Commun, 14, 2625.
- Dauparas, J., Anishchenko, I., Bennett, N., et al. (2022). Robust deep learning–based protein sequence design using ProteinMPNN. Science, 378(6615), 49–56.
- Jumper, J., Evans, R., Pritzel, A., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583–589.
- Eastman, P., Swails, J., Chodera, J.D., et al. (2017). OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLOS Computational Biology, 13(7), e1005659.
- Xue, L.C., Rodrigues, J.P., Kastritis, P.L., Bonvin, A.M.J.J., & Vangone, A. (2016). PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics, 32(23), 3676–3678.